Supervisors
Authors

Thakshajini Suhumar

Kapilrajh Ravindrakumar





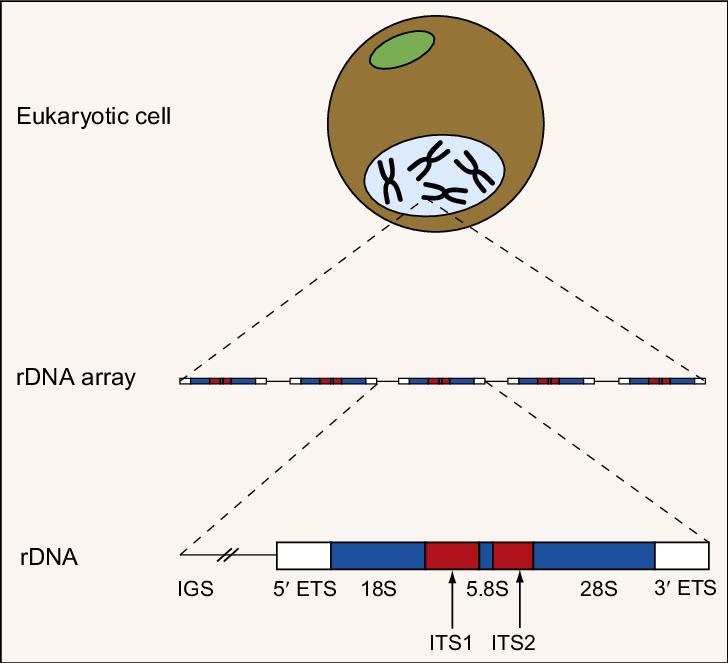
Internal Transcribed Spacer (ITS) region is vastly preferred among all genomic regions for phylogenetic studies associated with various plant species. Isolation of ITS regions from skim sequencing data results in more accurate inter-species as well as intra-species diversity analysis. Most of the previous studies utilized the available tools and pipelines to isolate fungal ITS regions from Illumina sequences. Botanists find it much difficult to gather plant ITS regions from skim sequencing data due to the lack of an efficient existing workflow. Our study focuses to come up with a workflow that comprises a user-friendly pipeline for the botanists to isolate plant ITS regions from Illumina skim sequencing data as accurately and as efficiently as possible for phylogenetic studies.