Supervisors
Authors

Kithma Madhushani
Nipuni Muthucumarana





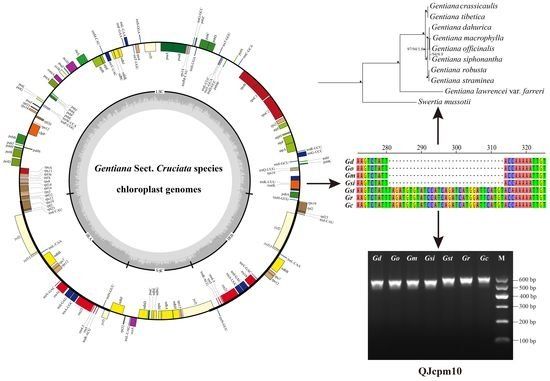
Chloroplast genes and genomes play an important role in plant phylogeny and species identification. Skim sequencing is getting low coverage genome sequencing data that has nuclear, chloroplast and mitochondria genome sequences. Since the fast development of high throughput sequencing technologies, it’s low cost to urge the low coverage data of the whole genome (usually concerning 20-30GB data), that is enough to assemble a whole chloroplast genome. To date, there are several assembly processes/ pipelines designed to assemble a whole chloroplast genome. However, what proportion knowledge is required or really utilized in such analysis is a problem. Having such information can facilitate biologists to style their experiments properly and cost-effectively. Biologists expect a straightforward, quick and easy procedure to assemble and annotate a circular chloroplast genome from Illumina NGS data. In this project, we’ll analysis the present procedures for chloroplast genome assembly and annotation, and work on developing the strategies to spot and choose the best set(s) of data and the procedure(s) to assemble a given chloroplast genome as accurately and efficiently, by statistical, computational and heuristic strategies.